Mutations at the interface of a protein-protein complex can alter the binding affinity. In the case of additive multiple mutations, the sum of the changes in binding free energy (ΔΔGbind) of individual mutations is approximately equal to the ΔΔGbind value of the complex containing all the mutations. Addem uses machine learning to predict whether a given pair of interface mutations are additive.
- When the user gives a particular PDB ID, Addem checks whether the given structure is that of a heterodimeric complex which does not contain nucleic acid (DNA/RNA).
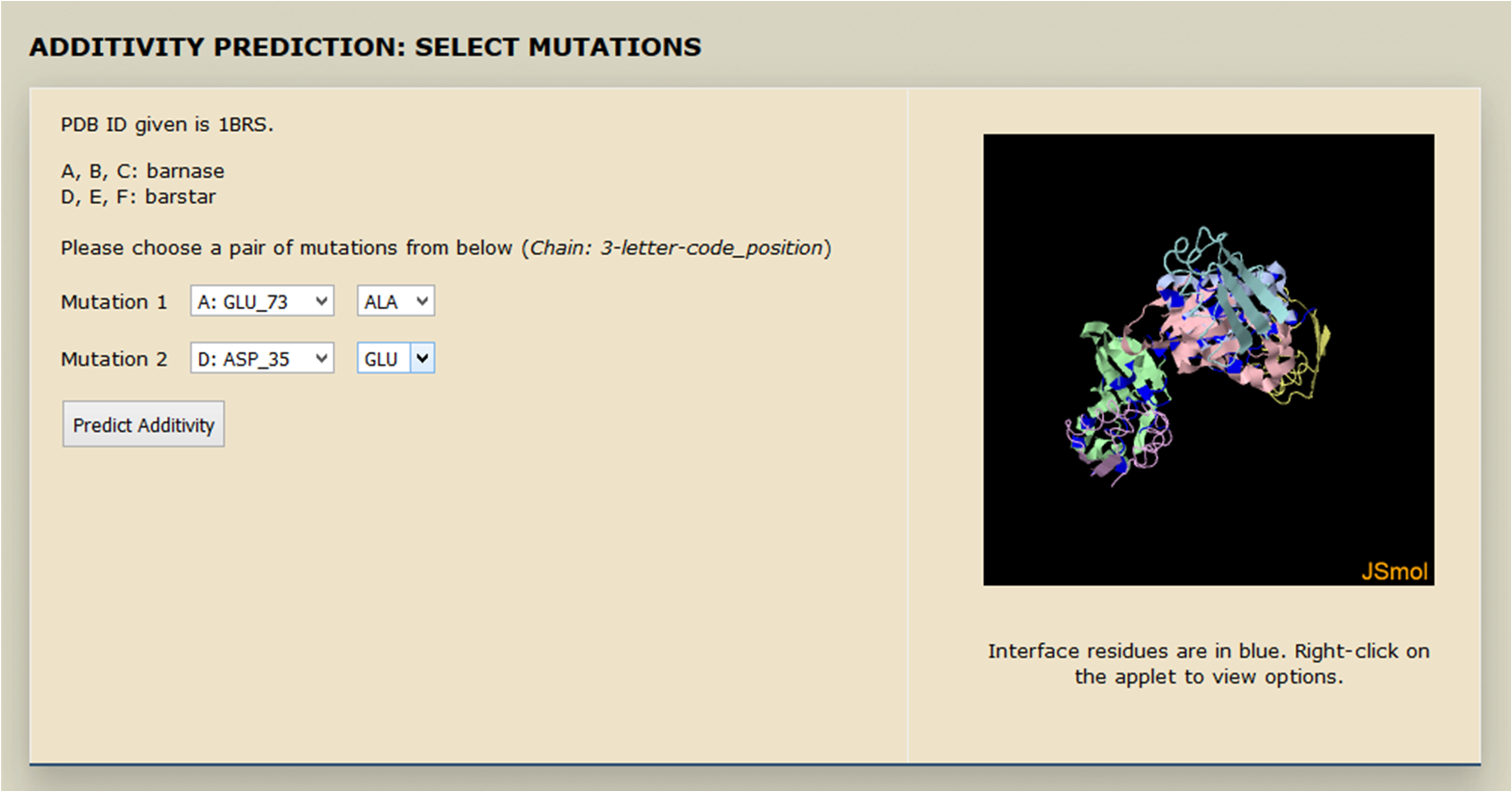
- If the complex structure is validated, Addem provides a list of interface residues. The user can refer to the JSmol applet for a view of the interface residues, which are displayed in blue.
- Once the user provides the mutations, Addem uses the Random Forest module from scikit-learn (machine learning package from Python) to predict whether the mutations are additive.
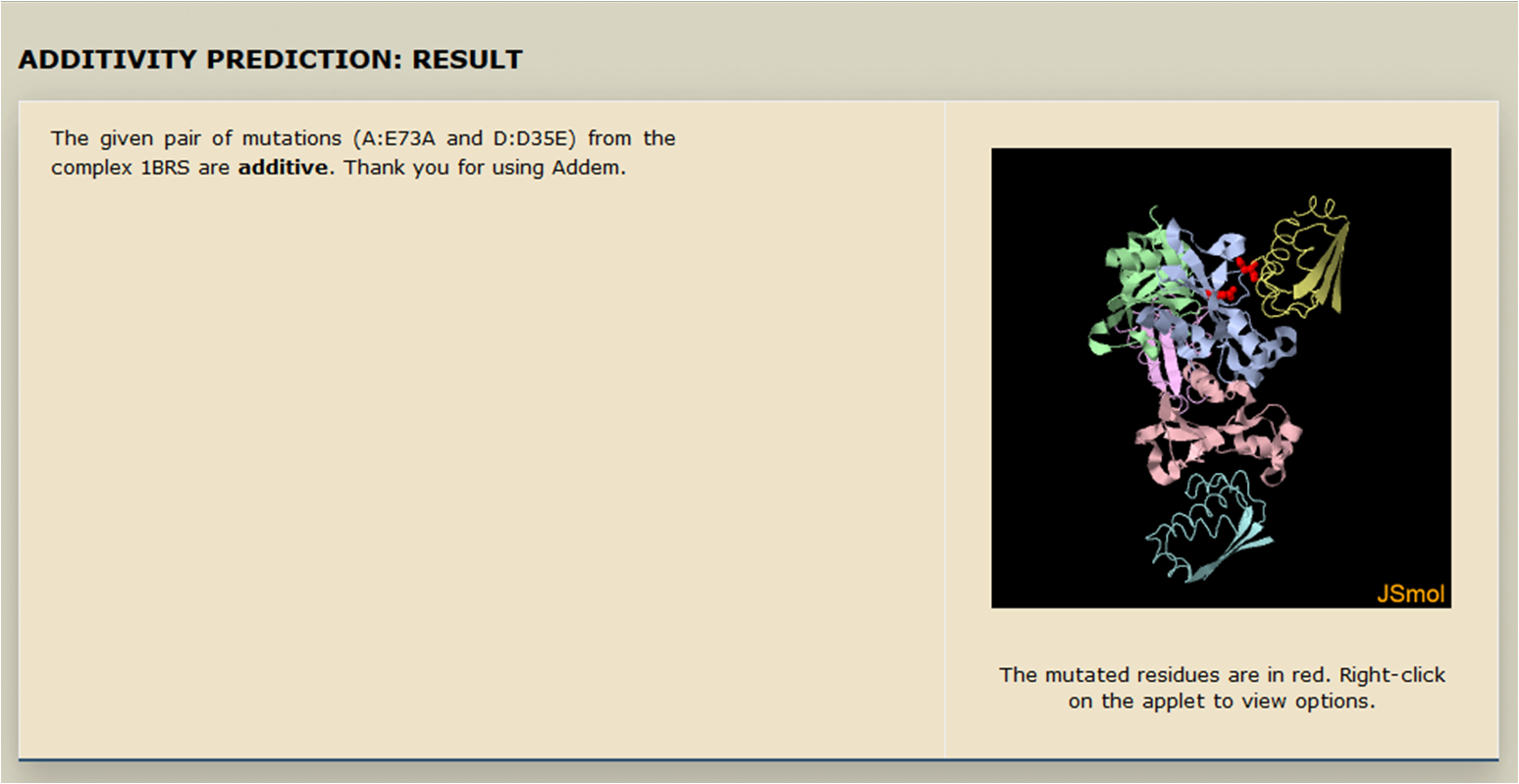
- The Result (additive or non-additive) is displayed along with visuals of the complex structure and the mutations.
- If the given pair of mutations are already available in PROXiMATE, the ΔΔGbind values are displayed for the single and double mutants, and whether it is additive or not.
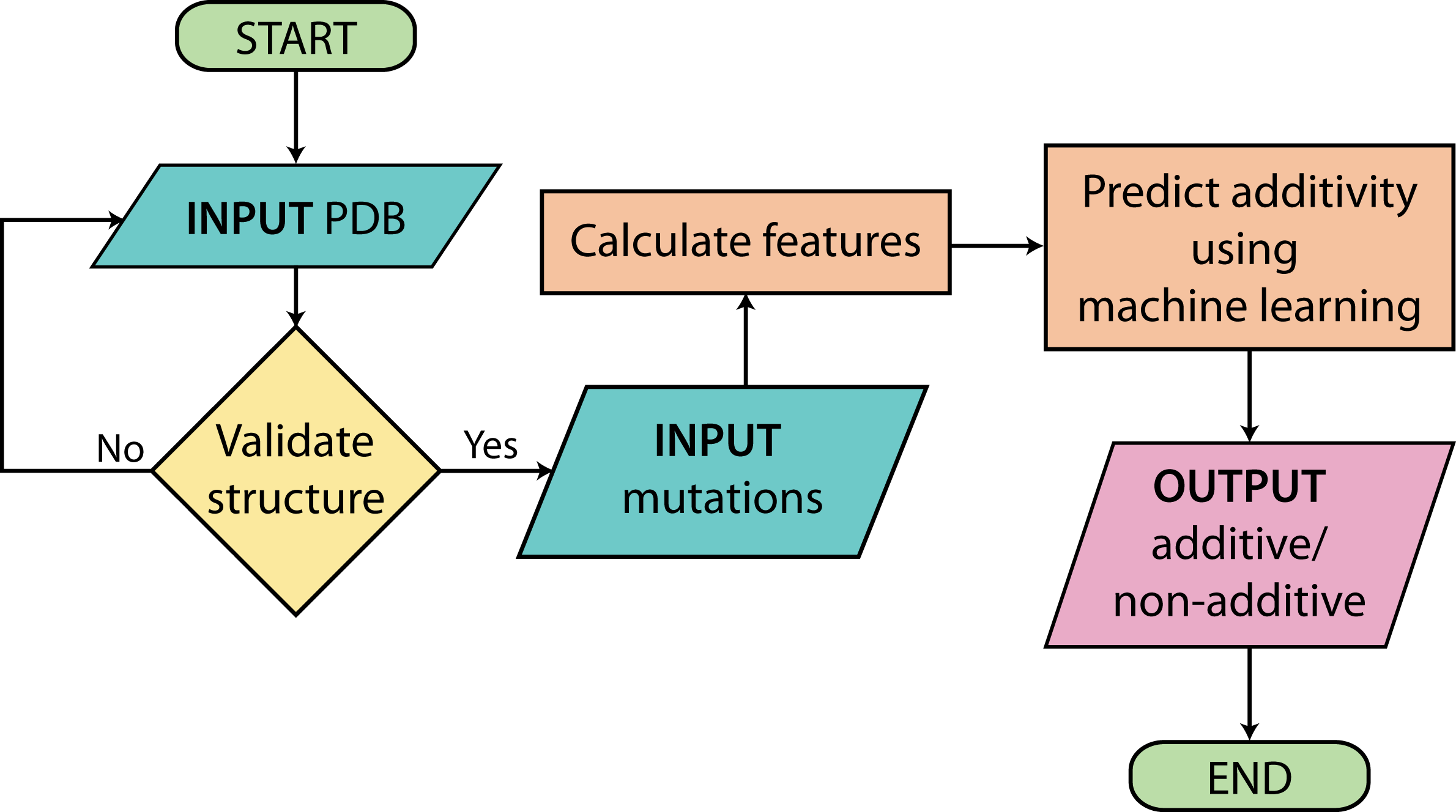 Flowchart outlining how Addem works
Flowchart outlining how Addem works
What is Addem?
Addem is a prediction server to predict whether the given pair of interface mutations in a given protein-protein complex are additive in terms of free energy of binding. This is NOT a prediction server for binding affinity of mutant protein-protein complexes. To know more about the concept of additivity of binding free energy, please see the questions below.
What are protein-protein interactions?
Nearly every cellular process (cell signalling, cell-cell adhesion, transport, metabolism, ubiquitination etc.) involves protein-protein interactions,
i.e. interactions between two or more protein molecules, usually mediated by various biophysical forces (such as electrostatics, Van der Waals forces,
hydrogen bonds, hydrophobic interactions etc.). The interaction is characterized by the formation of a protein-protein complex. For detailed reviews of protein-protein interactions, see the following references.
- Jones,S. and Thornton,J.M. (1996) Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA., 93(1), 13-20.
- Stites,W.E. (1997) Protein-Protein Interactions: Interface Structure, Binding Thermodynamics, and Mutational Analysis. Chem. Rev., 97(5), 1233-1250.
- Ali,M.H. and Imperiali,B. (2005) Protein oligomerization: How and why. Bioorgan. Med. Chem., 13(17), 5013-5020.
- Keskin,O. et. al. (2008) Principles of Protein-Protein Interactions: What are the Preferred Ways For Proteins To Interact? Chem. Rev., 108(4), 1225-1244.
- Janin,J. (2009) Basic Principles of Protein-Protein Interaction. In R. Nussinov and G. Schreiber (Eds.), Computational Protein-Protein Interactions., 1-19, Boca Raton: CRC Press.
What is additivity of binding free energy?
Additivity in binding affinity of protein-protein complexes refers to the change in free energy of binding (ΔΔGbind, in kcal/mol) for double (or multiple) mutations which is approximately equal to the sum of their corresponding single mutation ΔΔGbind values.
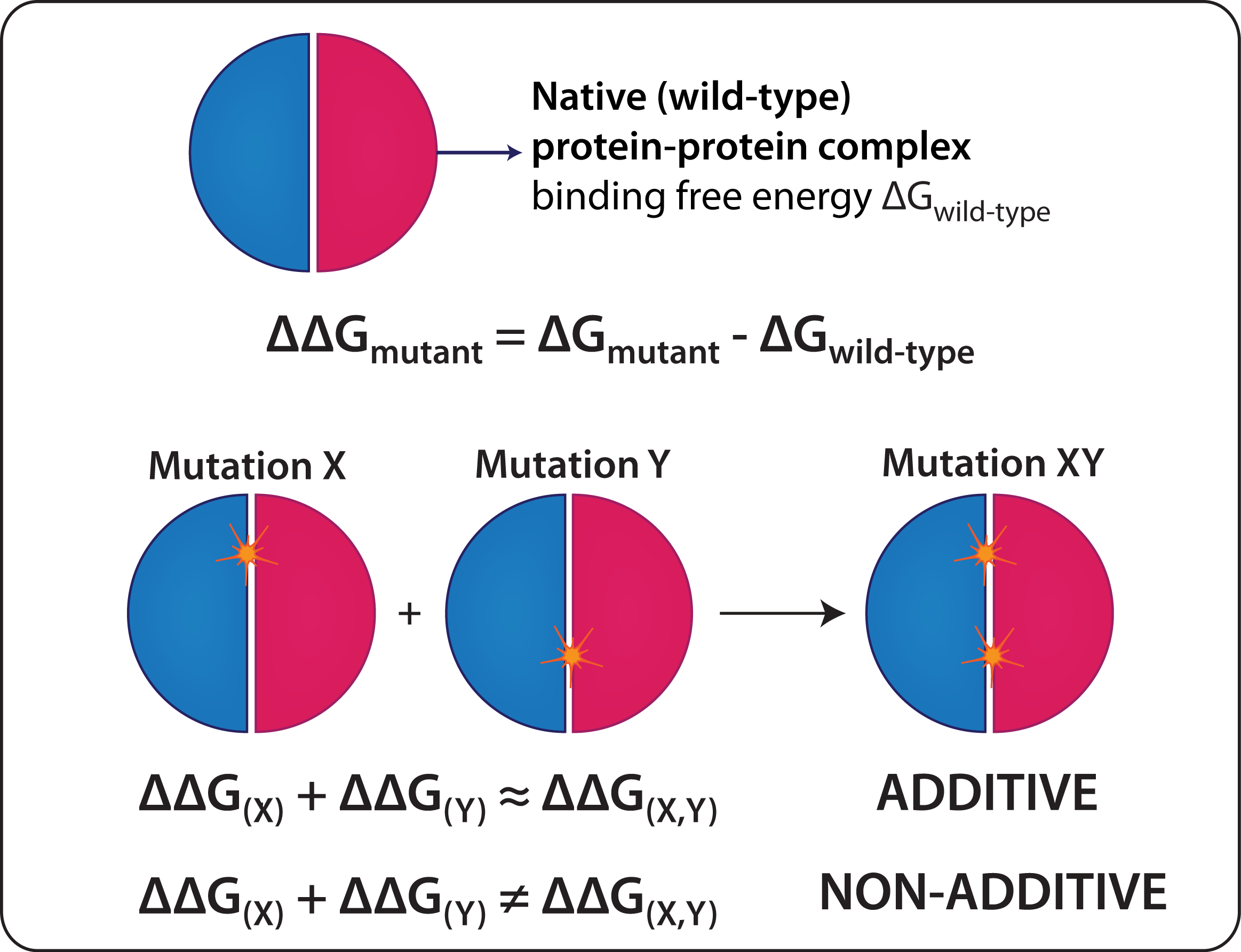 Additivity of binding free energy in protein-protein complexes
Additivity of binding free energy in protein-protein complexes
I am facing issues with the display of the website on my browser.
The website may be best viewed using the latest version of a browser such as IE10+, Google Chrome, or Mozilla Firefox.
I would like to know more about the concept of additivity.
Few studies have been done to study the additivity of binding free energy for mutant protein-protein complexes, for specific complexes or a small dataset of mutations from a few complexes (for e.g., Wells, 1990). Our study of additivity with a dataset of 379 double mutants has revealed significant insights, and the paper is under review.
I have some suggestions/feedback. How to reach you?
Please contact us by filling the form in the Contact page. You will receive a response shortly.