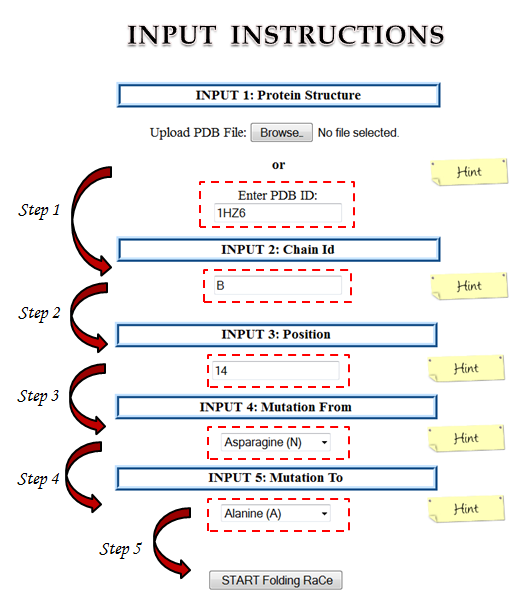
STEP 1:
(a) Click on upload tab to browse the protein coordinate text file in PDB format
or
(b) Mention the PDB ID in the space provided.
If both options are used together, then only the PDB Id will be processed and PDB uploaded by the user will be discarded.
STEP 2:
Mention the protein chain identifier for which the prediction is to made, even if your protein has a single chain.
In case of proteins with multiple chains or domains, folding rate change will be calculated only for individual chains in the proteins.
STEP 3:
Mention the position of residue to be mutated as provided in PDB file.
At times negative positions can also be mentioned in the PDB file.
Mention as it is along with negative sign. Server will take care of it.
STEP 4:
Select the wild residue from the drop down menu at the above mentioned position, as per the PDB file.
STEP 5:
Select the desired mutant residue from the drop down menu whose effect on folding is needed to be calculated.
Please Note: All fields are mandatory.
Mouse-over the "Hints" for brief description of each section in input.
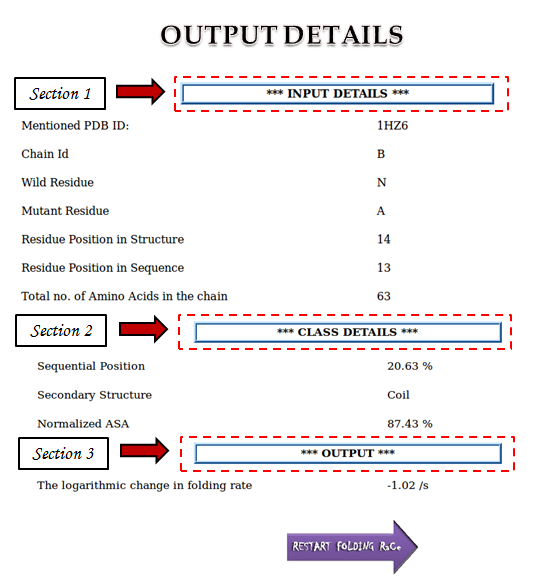
Displays the details of the input file as well as other mandatory fields to be filled by user.
Self Explanatory Errors will be displayed if the uploaded file is not complying with the specified format or corrupt or exceeds 200 Mb as well as if the mentioned PDB ID does'nt exist.
Such errors will help the user to troubleshoot any mistakes while entering the input details.
Section 2: CLASS DETAILS
If the input details are correct, then server will proceed to the next section, wherein, it will specify the three classes of the mutant position viz. sequential position, secondary structure and accessible surface area.
Section 3: OUTPUT DETAILS
This section provides the predicted value of folding rate change upon specified mutation in the given protein in logarithmic scale.